Camila Farias Amorim, Ph.D.
Immunology - Clinical/Translational Medicine
Computational Biology - Bioinformatics
OMICS - Molecular Biology
About Me
Hello! Welcome to my website.
Computational Biology & Immunology PhD scientist specialized working in Omics, molecular profiling, and AI-enabled biomarker discovery to support allogeneic iPSC-derived cell therapies, spanning immune (CAR-T/CAR-NK) and non-immune (SC–derived pancreatic islet) programs for oncology and Type 1 Diabetes.
Bringing 12+ years of experience across wet- and dry-lab settings, integrating cellular biology, immunopathogenesis, and high-dimensional genomics to enable mechanism-based biomarker discovery, ML predictive modeling, and translational decision-making across preclinical and early clinical development.
Incorporating AI-assisted analytics, including large language models (LLMs), to support workflow automation, reproducible analysis, scalable data exploration, and hypothesis generation in biomarker and target discovery.
Current role
Currently working on a Principal Scientist role at Century Therapeutics Inc., LinkedIn
Leading the NGS and molecular assay strategies across research, cell therapy manufacturing, and external stakeholder communication. Scope includes study design, assay development (qPCR, flow cytometry, CITE-seq, scRNA-seq, spatial transcriptomics), PK/PD and statistical modeling, and multi-omics data integration to inform product optimization and program strategy.
Working horizontally across iPSC biology, immunology, differentiation, translational, manufacturing, and clinical teams, and oversees Omics-focused collaborations with CROs and technology vendors, ensuring experimental rigor, data quality, and cross-study comparability.
I lead company-wide Research Omics forums to align wet- and dry-lab teams and translate complex NGS datasets into actionable insights. Background includes biomarker and target discovery, predictive modeling, immunotherapies, vaccines, and early clinical biomarker strategy, with prior experience teaching NGS and bioinformatics at the University of Pennsylvania and 20+ peer-reviewed publications.
Focusing on connecting biological insight, quantitative analysis, and collaborative execution to support rigorous science and patient-relevant outcomes.
Avaliable materials:
1. Omics lead in: iPSC-derived CD4+, CD8+ αβT cells that expand upon antigen stimulation and exhibit in vivo tumor control. 2025. Keystone poster
#ComputationalBiology #BiomarkerDiscovery #PredictiveModeling #MolecularProfiling #LLMs #SingleCell #SpatialTranscriptomics #CellTherapy #iPSC #CAR_T #CAR_NK #SCIslets #TranslationalScience #Omics #NGS #CROManagement #DEI
Keywords
high-throughput NGS; AI/LLMs; ML predictive modeling; R; python; C++; bash; Unix/Linux OS; command-line tools; shell scripting; read mapping; Illumina sequencing; DNA; RNA; RNAseq, Dual-RNAseq; single-cell RNAseq, CITE-Seq, 16S-seq, WGS, clinical metadata; Bioconductor; tidyverse; data visualization; linear modeling; machine learning; supervised/unsupervised classification; hierarchical clustering; dimensionality reduction; pattern recognition; git/GitHub; version control; cloud; ShinyApp; datasets; ggplot; statistical inference; pathway analysis; GSEA; GO; GEO NCBI
Academic publications
A complete list of publications can be found in my Google Scholar Profile or more on Web of Science. See below my main published projects:
Variable gene expression and parasite load predict treatment outcome in cutaneous leishmaniasis
 Biological significance: Most of the patients infected by L. braziliensis fail the therapy with an anti-parasitic drug, and there are no vaccines for this infection.
I discovered that both parasite burden and the cytotoxic effector mechanism from skin CD8+ T cells/NK cells affect the response to therapy and can be used as an early biomarker at the clinic.
The immunotherapy targeting this later inflammatory mechanism in experimental models dramatically impacted the control of the disease.
Biological significance: Most of the patients infected by L. braziliensis fail the therapy with an anti-parasitic drug, and there are no vaccines for this infection.
I discovered that both parasite burden and the cytotoxic effector mechanism from skin CD8+ T cells/NK cells affect the response to therapy and can be used as an early biomarker at the clinic.
The immunotherapy targeting this later inflammatory mechanism in experimental models dramatically impacted the control of the disease.
Bioinformatics: This was my first publication merging Immunology and Computational Biology. I investigated a human skin, lesional RNA-seq dataset from Brazilian cutaneous leishmaniasis patients and developed a workflow focused on genes with a highly variable expression between samples. In this study, R statistical programming language, dual-RNAseq mapping (quantifying both human and parasite reads from the samples), and a combination of strategies to reduce the dimensionality of the metadata were used to find these genetic biomarkers. All the code involved in reproducing the analysis and images is available in a CodeOcean capsule.
Farias Amorim et al. 2019, Science Translational Medicine, DOI: 10.1126/scitranslmed.aax4204
Code capsule: CodeOcean
Multi-omic profiling of cutaneous leishmaniasis infections reveals microbiota-driven mechanisms underlying disease severity
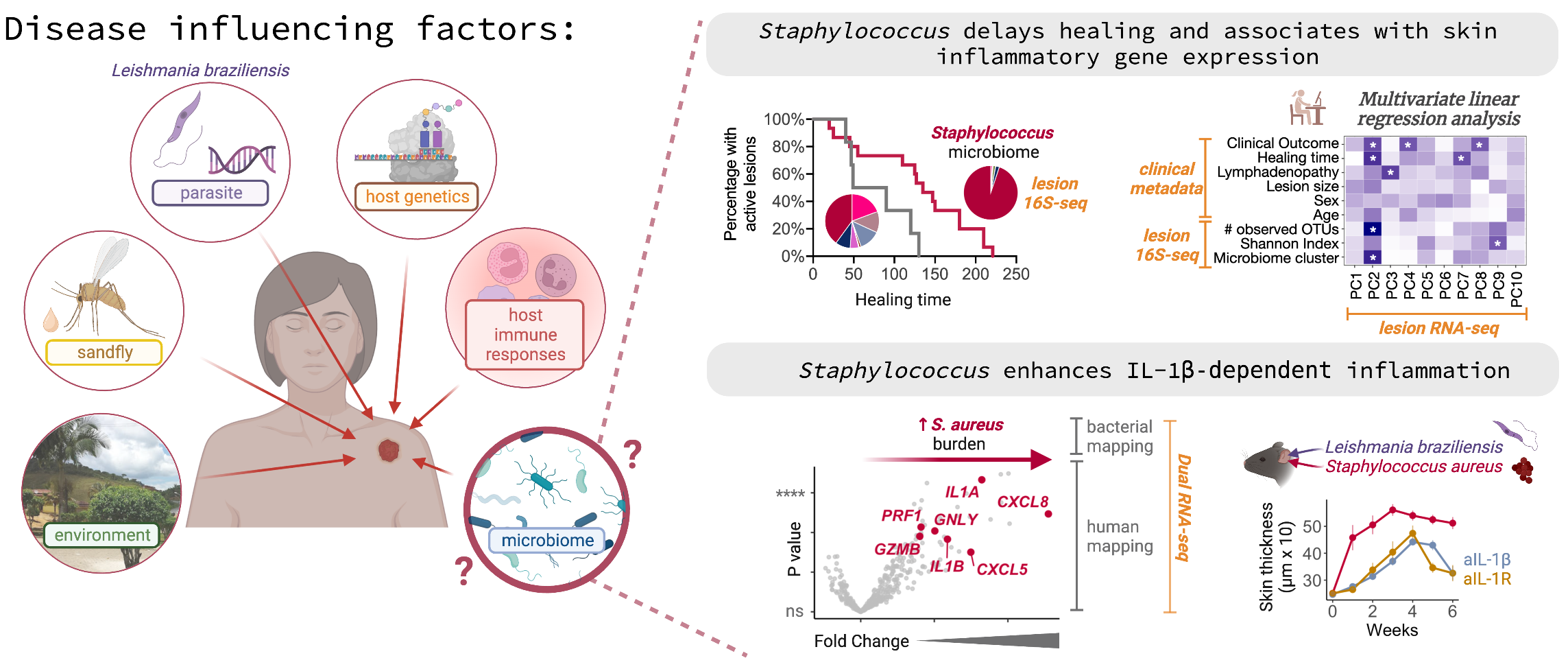 Biological significance: The development of the lesion caused by the Leishmania braziliensis infection in humans is influenced by several factors.
I collaborated with a big team of Brazilian clinicians and experimentalist bench scientists and analyzed a multi-omics dataset from those patients.
We discovered that the skin microbiome strongly contributes to the severity of the disease, delayed healing, and unrevealed IL-1b as a major inflammatory mediator behind this association.
This study's results support targeted immunotherapies and bacterial therapies for this parasitic infection.
Biological significance: The development of the lesion caused by the Leishmania braziliensis infection in humans is influenced by several factors.
I collaborated with a big team of Brazilian clinicians and experimentalist bench scientists and analyzed a multi-omics dataset from those patients.
We discovered that the skin microbiome strongly contributes to the severity of the disease, delayed healing, and unrevealed IL-1b as a major inflammatory mediator behind this association.
This study's results support targeted immunotherapies and bacterial therapies for this parasitic infection.
Bioinformatics: I have reduced the dimensionality, integrated and analyzed a multi-omics dataset of L. braziliensis patients (skin RNA-seq, 16S-seq, bacterial WGS, qPCRs, and clinical metadata). I developed a computational workflow with the R programming language based on multivariate linear regression analysis and machine learning methods to identify elements contributing to the worse clinical outcome.
Farias Amorim et al. 2023, Science Translational Medicine, DOI: 10.1101/2023.02.02.23285247
Localized skin inflammation during cutaneous leishmaniasis drives a chronic, systemic IFN-γ signature
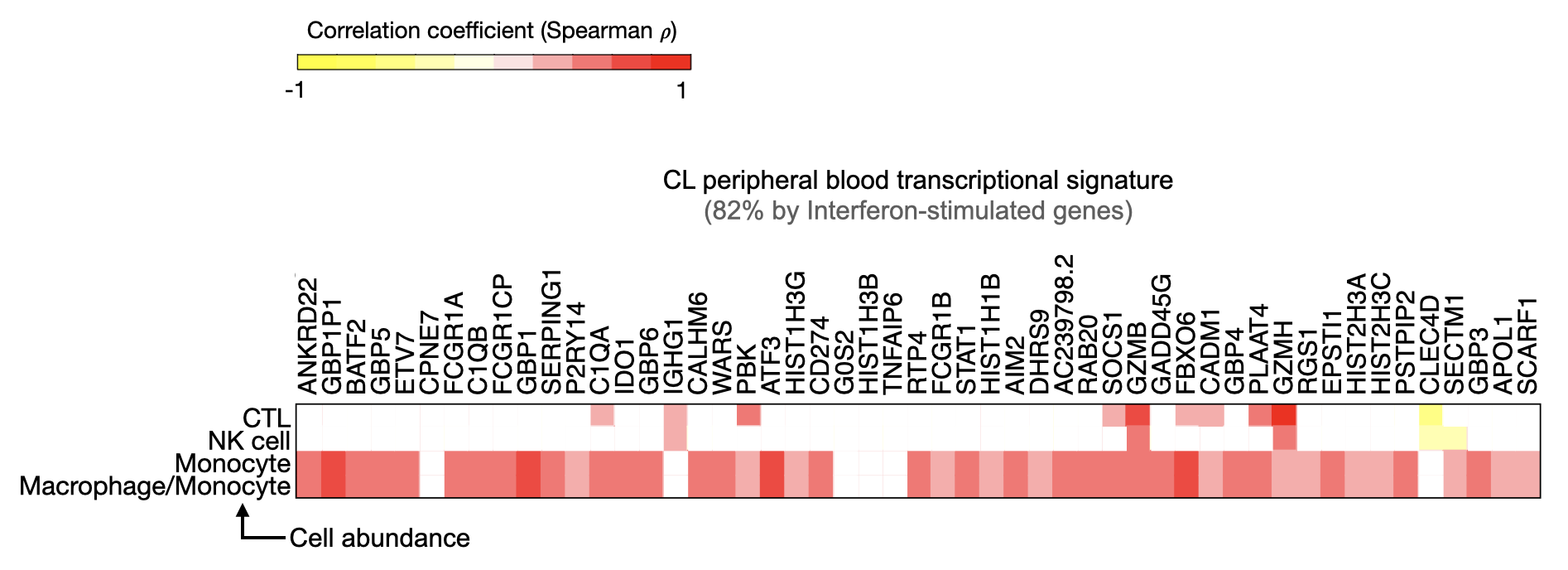 Biological significance: In this study, we were interested in evaluating whether the localized skin infection caused by the Leishmania braziliensis impacts patients' systemic immunity.
We discovered a monocyte-Interferon-gamma and CD8+ T cells cytotoxic (CTL) transcriptional inflammation at the site of skin infection and systemically (lesion and peripheral blood RNA-seq analysis).
Moreover, when we compared the blood from those patients with subjects with other diseases (tuberculosis and malaria), we identified the transcriptional CTL signature as a unique circulatory feature of this parasitic disease.
We amplified our knowledge about this localized skin condition: cutaneous leishmaniasis. We raised the question of how these systemic responses might influence the response to other infections and the overall health of these individuals.
Biological significance: In this study, we were interested in evaluating whether the localized skin infection caused by the Leishmania braziliensis impacts patients' systemic immunity.
We discovered a monocyte-Interferon-gamma and CD8+ T cells cytotoxic (CTL) transcriptional inflammation at the site of skin infection and systemically (lesion and peripheral blood RNA-seq analysis).
Moreover, when we compared the blood from those patients with subjects with other diseases (tuberculosis and malaria), we identified the transcriptional CTL signature as a unique circulatory feature of this parasitic disease.
We amplified our knowledge about this localized skin condition: cutaneous leishmaniasis. We raised the question of how these systemic responses might influence the response to other infections and the overall health of these individuals.
Bioinformatics: I used linear modeling, statistical inference, and data visualization strategies to profile the systemic transcriptome of patients with cutaneous leishmaniasis and revealed the gene expression patterns associated with clinical variables and outcomes. I also downloaded publicly available blood RNA-seq datasets from patients with other infections and compared their transcriptomes to identify disease-specific data patterns.
Farias Amorim et al. 2021, PLoS NTDs, DOI: 10.1371/journal.pntd.0009321
Teaching: An Open-Source Toolkit To Expand Bioinformatics Training in Infectious Diseases
 I was a Teaching Assistant in the DIYtranscriptomics.com course led by Dr. Beiting at the University of Pennsylvania.
As a short description (almost unfair), we teach how to work with transcriptomic data (bulk and single-cell RNA-seq): from installing the necessary tools to writing base/tidy R, using Bioconductor R packages, data modeling, and final next-generation sequencing interpretation of biological results.
We use examples of real-world datasets in the context of Infectious Diseases to introduce updated multi-omics concepts from the greater area of Computational Biology.
The course is fully available online, and our team put together a paper gathering everything we have learned in past years.
I was a Teaching Assistant in the DIYtranscriptomics.com course led by Dr. Beiting at the University of Pennsylvania.
As a short description (almost unfair), we teach how to work with transcriptomic data (bulk and single-cell RNA-seq): from installing the necessary tools to writing base/tidy R, using Bioconductor R packages, data modeling, and final next-generation sequencing interpretation of biological results.
We use examples of real-world datasets in the context of Infectious Diseases to introduce updated multi-omics concepts from the greater area of Computational Biology.
The course is fully available online, and our team put together a paper gathering everything we have learned in past years.
Berry and Farias Amorim et al. 2021, mBio, DOI: 10.1128/mBio.01214-21
Code capsule: CodeOcean
